A Research Article Digest: Short Range Mutational Biases Largely Define Genome Heptameric Composition
Research Article Title: Single genome retrieval of context-dependent variability in mutation rates for human germline.
Journal: BMC Genomics, 2017, 18, 81.
http://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-016-3440-5
(first deposited in bioRxiv on 10 August 2015)
Simple Short Explanation of the Work for Everyone:
Journal: BMC Genomics, 2017, 18, 81.
http://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-016-3440-5
(first deposited in bioRxiv on 10 August 2015)
Simple Short Explanation of the Work for Everyone:
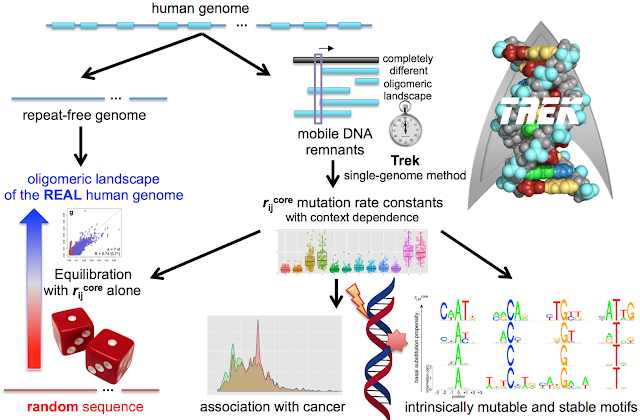
Our genome is a very long DNA strand holding a sequence of nucleobases. Those nucleobases are of four types - letters A, T, G and C - and there are ~3.5 billion such letters in our genome. However, the letters do not stay the same over evolutionary timescale (millions to billions of years), and very slowly change/swap (or, as we call it, mutate) in our genome, driving the evolution of life. The work presents a technique to retrieve the speed with which those letters change/swap by accounting the full extent on how the neighbouring letters tune the speed. We use only a single genome, rather than requiring thousands of genomes as needed for such an undertaking, and our single-genome approach is important in eliminating some biases due to the differences in the speeds of mutations from one individual to another. The work independently demonstrates that, in our DNA, 7-letter “words” or chunks are what define how speedy or slow the mutations are.
Most importantly, the work shows that the composition of our genome, if expressed by the content of such (up to) 7-letter words, is prevalently determined by the sequence-context biases in the speeds of mutations, rather than other higher effects. This means that some DNA words, such as GAACAGT, are less frequent in our genome, and some are more frequent, due to the increased or decreased intrinsic speed with which they change in time. In particular, when we take a completely random sequence of letters and “evolve” it inside our computers by using only our found speed numbers, we arrive into a state (a simulated genome) with its nucleobase content and 16,384 (4^7) 7-letter word/chunk frequencies mirroring that of the actual real human genome. This therefore shows that seemingly complex compositional (up-to 7-letter) patterns in genomes originate from fundamental simpler properties, just like protein 3D global structures are fully defined by fundamental, shorter-range forces.
We also directly demonstrate that cancer is mostly associated with mutations that happen in less-stable DNA chunks, and envision the usage of our outcomes to shed light on the evolution and stability of our genomes.


Comments
Post a Comment